By
Kevin E. Noonan —
A
generation ago, Jeremy Rifkin famously convinced the Cambridge city council to
ban genetic engineering in that city, using the fear of "tinkering"
with nature and producing a "superbug" that would hurt the public
health and thereby (for a time) reduced progress in biological research at
Harvard and elsewhere in the city environs. Mr. Rifkin was not alone in his
concern over potential problems from recombinant DNA; indeed, the scientist who
first produced a recombinant plasmid, Paul Berg (Jackson et al.,
1972, "Biochemical Method for Inserting New Genetic Information into DNA
of Simian Virus 40: Circular SV40 DNA Molecules Containing Lambda Phage Genes
and the Galactose Operon of Escherichia coli," Proc.
Natl. Acad. Sci. U.S.A. 69(10): 2904-09),
was instrumental in organizing a conference in 1975 on the issue at Asilomar,
California, which resulted in guidelines (both physical and biological) from
the National Institutes of Health for the safe practice of the new technology (Guidelines
for research involving recombinant DNA molecules," 41 Fed. Reg. 27911-43 (1976)). But in
the intervening decades, widespread distribution of laboratory-derived recombinant
DNA introduced into the environment has not been reported as a threat to humans
or the environment.
 Until
Until
now, that is. In a report in the journal
Environmental Science and Technology
entitled "A Survey or Drug Resistance bla
genes Originating from Synthetic Plasmid Vectors in Six Chinese Rivers,"
scientists from Sichuan University and the Chinese Institute of Health and
Environmental Medicine report that drug resistance genes (specifically, the
beta-lactamase (bla) gene that
provides resistance to ampicillin) have been detected in the Pearl, Yangtze,
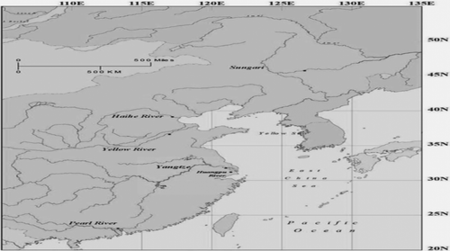
Yellow, Hai He, Sunggari, and Huangpu rivers in mainland China (Chen et al., 2012, Environ. Sci. Technol. 46(24): 13448–54). These rivers drain the majority of the
Chinese mainland in all provinces and have the cities of Guangzhou, Nanjing, Ji'nan,
Tianjin, Harbin, and Shanghai, respectively, along their banks. The authors took samples from these rivers
containing naturally occurring bacteria and assayed using polymerase chain
reaction (PCR) and a variant, quantitative real-time PCR, to assess the
frequency with which beta-lactamase DNA could be detected. The assay was specific for the bla gene that comprises most recombinant
plasmid strains, such as pBR322 (one of the oldest and most widespread plasmids
used for molecular biological research) as well as pUC19. PCR was performed using three pairs of "universal"
primers for the bla gene as well as
sequencing primers used for detecting exogenous DNA in cloning sites in the
plasmid vectors. In addition, the
bacterial isolates obtained from the river samples were assayed for antibiotic
resistance using assays recommended by the U.S. Clinical Laboratory Standards
Institute.
Plasmid-derived
bla-encoding DNA was detected in all
six rivers assayed, detected using both the "universal" primers as
well as the plasmid sequencing primers (the latter results further supporting
the conclusion that the samples were of laboratory origin). The detection rate varied from 21.9% (in the
Hai He River samples) to 36.4% (in the Yangtze River samples), and the total
copy number of bla copies in the
samples ranged from 6.7 +/- 5.0 x 101 copies/mL (in the Yellow River
samples) to 4.6 +/- 0.6 x 103 copies/mL (in the Pearl River
samples). The Pearl and Hai He rivers
showed the widest spectrum of cephalosporin resistance from the bla gene present in bacterial samples,
extending to 3rd- and 4th-generation drugs like
cefotaxime and cefoperazone, while the spectrum was narrower (e.g., cefalotin, cephazolin,
cefmetazole, and cefoxitin) in samples from the other rivers tested. Sequence analysis confirmed that sequences "neighboring"
the bla sequences detected in the
river samples "most frequently represented artificial or synthetic
constructs, including cloning, expression, shuttle, gene-fusion, and gene trap
vectors" derived from recombinant laboratory plasmid vectors, most
strongly with pBR322.
An
interesting ancillary result reported in the paper was that tetracycline
resistance-conferring DNA was also detected in samples from all six rivers, and
some river samples also showed genes that provided resistance to other commonly
used laboratory antibiotic selection markers such as gentamicin. Detection of gene sequences encoding
tetracycline resistance is consistent with detection of pBR322, which was
engineered to contain a tetracycline resistance gene as well as the bla gene. The authors also report that detection of
high levels of laboratory plasmid-derived antibiotic resistance genes in the
Pearl River was consistent with the levels of antibiotic pollution know to
exist in that river from human and animal sources.
This
report raises serious questions, of course, over the consequences of widespread
use of antibiotic resistance-encoding genetic elements. It is particularly disturbing because genetic
engineering is not limited to the lab anymore, being used in such technologies
as biofuels, agriculture, and bioremediation. The risk of antibiotic resistance gene contamination of the environment
has been recognized previously and steps taken to minimize it; for example,
Monsanto has developed vectors encoding other markers such as lacZY for use in its recombinant seed. These efforts take on greater significance,
and relevance for human and animal well-being, in view of this latest report.

Leave a comment