By Kevin E. Noonan —
 The recent discussion of the Federal Circuit's decision in the In re Kubin case suggests there may be some misunderstanding of the science behind the legal question of obviousness. Because obviousness is a question of law based heavily on underlying issues of fact, it is important for commentators as well as judges to have some understanding of the underlying science. The Federal Circuit's decision in the Kubin case illustrates the possible consequences of misunderstanding, or of Judge Rich's recitation of the "rudiments" of biotechnology from In re O'Farrell as somehow conferring on appellate judges the illusion that their understanding is per se sufficient.
The recent discussion of the Federal Circuit's decision in the In re Kubin case suggests there may be some misunderstanding of the science behind the legal question of obviousness. Because obviousness is a question of law based heavily on underlying issues of fact, it is important for commentators as well as judges to have some understanding of the underlying science. The Federal Circuit's decision in the Kubin case illustrates the possible consequences of misunderstanding, or of Judge Rich's recitation of the "rudiments" of biotechnology from In re O'Farrell as somehow conferring on appellate judges the illusion that their understanding is per se sufficient.
We must start, generally and classically, with a protein. (The complications involved with the massive amount of sequence information generated by the Human Genome Project will be discussed separately below.) Proteins, and the extent of knowledge in the prior art about proteins, varies enormously, from simple knowledge of its existence (as in Kubin) to a complete determination of the amino acid sequence of the protein, as in In re Bell. Knowledge commensurate with Bell limits the scope of any claim for a gene encoding the protein to the specific nucleotide sequence of the isolated cDNA. This is because of the degeneracy of the genetic code: there are multiple codons for all but two of the 20 naturally-occurring amino acids, so the number of possible nucleotide sequences for any amino acid sequence is typically astronomical — the number of possible nucleotide sequences that could encode the insulin-like growth factor in the Bell case was 1036 (for a protein encoded by 70 amino acids). Non-obviousness on these grounds is Federal Circuit precedent from the Bell case, and neither Kubin nor KSR International Co. v. Teleflex Inc. overrules it (unless 1036 can be considered a "small" number of predicable solutions).
More typically, only partial amino acid sequence information from a protein is known in the art, of variable reliability (albeit the reliability has improved over the past 25 years). This knowledge is used to produce probes for screening libraries prepared according to the Sambrook reference, but the predictability of being able to isolate a cDNA encoding the desired protein depends on whether the amino acid sequence known in the art can produce a degenerate probe that reliably detects a cDNA clone in the library. Alternatively, longer probes can be developed depending on conserved functional motifs in the encoded protein (such as ATP-binding cassettes or G-protein coupled receptors). However, the efficiency of using such probes depends on how closely conserved the particular motif is in the protein of interest, which is frequently not known in the prior art. (As in Kubin, there are also ways of screening that rely on expression of antigenic epitopes of the protein of interest that are detected by antibodies, but the principles are the same.)
 This screening step depends upon more that the quality of the probe, however. Most importantly, the art needs to provide a source for messenger RNA that can be used to make the cDNA library. In addition, the cell or tissue source must make enough of the specific mRNA to be cloned so that its representation in the library (i.e., the number of cDNAs produced from the specific mRNA) is high enough to be detected. This is because cells and tissues express mRNAs in generally broad classes of abundance: structural proteins like actin and tubulin are expressed at high abundance, while cell- or tissue-specific genes are frequently expressed at low abundance. While there may be some clues in the art about cell or tissue sources of an mRNA and the expression abundance thereof, these are generally gene-specific factual matters that cannot be presumed when making an obviousness determination. This is why Judge Rader (at left) was incorrect when he said "No, [Sambrook] tells one of skill in the art how to produce those libraries, and when you [have] the probe it's not so hard to do" during the Kubin oral argument, since Sambrook did not provide any teaching on what cells to use or how to treat them to stimulate production of NAIL-encoding mRNA to detectable levels.
This screening step depends upon more that the quality of the probe, however. Most importantly, the art needs to provide a source for messenger RNA that can be used to make the cDNA library. In addition, the cell or tissue source must make enough of the specific mRNA to be cloned so that its representation in the library (i.e., the number of cDNAs produced from the specific mRNA) is high enough to be detected. This is because cells and tissues express mRNAs in generally broad classes of abundance: structural proteins like actin and tubulin are expressed at high abundance, while cell- or tissue-specific genes are frequently expressed at low abundance. While there may be some clues in the art about cell or tissue sources of an mRNA and the expression abundance thereof, these are generally gene-specific factual matters that cannot be presumed when making an obviousness determination. This is why Judge Rader (at left) was incorrect when he said "No, [Sambrook] tells one of skill in the art how to produce those libraries, and when you [have] the probe it's not so hard to do" during the Kubin oral argument, since Sambrook did not provide any teaching on what cells to use or how to treat them to stimulate production of NAIL-encoding mRNA to detectable levels.
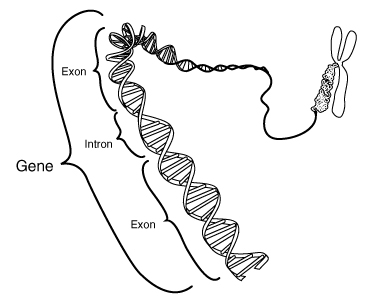 Now we come to the Human Genome Project (HGP), which has put lots of sequences into public databases and impacts both anticipation and obviousness. For anticipation, there will frequently be In re Hall issues raised: was the sequence sufficiently annotated so that the skilled worker would recognize it in the database? This is not a trivial question, due to one of the vagaries of genome structure (at least in most eukaryotic organisms like mammals and vertebrates). This feature is that genes are not set forth "in one piece" in the genomic DNA (which is the DNA sequenced in the Human Genome Project). Instead, the gene sequences encoding a protein are broken up by non-coding sequences (variably termed "intervening sequences," "introns" or "junk DNA"). This gives the typical gene the structure (see link): beginning of protein code — intron — continuing protein code — intron — end of protein code.
Now we come to the Human Genome Project (HGP), which has put lots of sequences into public databases and impacts both anticipation and obviousness. For anticipation, there will frequently be In re Hall issues raised: was the sequence sufficiently annotated so that the skilled worker would recognize it in the database? This is not a trivial question, due to one of the vagaries of genome structure (at least in most eukaryotic organisms like mammals and vertebrates). This feature is that genes are not set forth "in one piece" in the genomic DNA (which is the DNA sequenced in the Human Genome Project). Instead, the gene sequences encoding a protein are broken up by non-coding sequences (variably termed "intervening sequences," "introns" or "junk DNA"). This gives the typical gene the structure (see link): beginning of protein code — intron — continuing protein code — intron — end of protein code.
When the gene sequence is transcribed into RNA, at first the whole sequence, coding and introns alike are transcribed. Then, cellular enzymes splice out the introns, leaving the mRNA with the coding sequence arranged in one contiguous piece. cDNA is prepared from mRNA because it has the coding sequence in this linear fashion and has the least amount of "extra" RNA, usually at each end. This makes cDNA cloning much more efficient that genomic cloning.
But these aspects of cellular biology have consequences for what we can tell from the HGP data. First, is the data is present in the database in such a way that the skilled worker would be able to identify the "open reading frames" (i.e., portions of genomic DNA that could potentially be translated into protein via an mRNA intermediate) that encode the complete gene. This is not always the case, because a gene made up of 1,000 nucleotides may have introns that are 35,000 nucleotides in length. Certai
nly, as the data has been analyzed more and more vigorously these sequences have become more and more accessible and algorithms are available having the capacity to detect open reading frames separated by large intron sequences and to identify properly positioned splice sites that would predict how these open reading frames could be assembled into an mRNA encoding the desired protein. Thus, it has become more and more likely that the Hall conditions will be satisfied. But this will be a fact-specific question to be answered anew for each gene under consideration. For example, transcripts can be alternatively spliced in some instances and if the art did not disclose this the cDNA ultimately obtained would not be anticipated by the "predicted" sequence in the HGP data.
Returning to obviousness, one of the hallmark discoveries of the HGP is the detection of genes (and hence proteins) otherwise never before identified or suspected; this is particularly true of members of multigene families, especially those more distantly related by evolution. It seems that these genes should fall outside the Kubin standard of DNA obviousness because they lack the fundamental predicate of the decision: knowledge in the art that the protein existed in the first place. Indeed, the issue over the patentability of these genes has not been (and should not be after Kubin) whether they were obvious; the issue is whether applicants claiming these genes know (and disclose in their applications) the function of the encoded protein, its activity, in order to satisfy the utility requirements of 35 U.S.C. § 101. So it isn't as easy as looking up open reading frames in a database and filing a patent application for these "unknown" genes identified through the HGP.
It is fair to ask, if these are all the reasons why the Kubin sequence is non-obvious, under what circumstances could it be obvious? I think these factors would support an obviousness determination: the protein was known to exist in the prior art; the art identifies a cell or tissue type that expresses the gene of interest in amounts that permit cDNA from that gene to be represented in the library at levels (>1 clone in 100,000, for example) where the clones would be readily detectable (whether natively or as the result of some experimental treatment or manipulation); the art provides a probe (whether stemming from a reliable amino acid sequence encoded by a sufficiently non-degenerate collection of oligonucleotides or with a monoclonal antibody such as Valiante's C1.7 mAb) that efficiently detects the desired cDNA sequence; and the cDNA contained no features (hairpin structures, alternative splice sites, other anomalies) that would preclude or prevent cloning using conventional methods.
Dwight Eisenhower said "[f]arming looks mighty easy when your plow is a pencil and you're a thousand miles from the corn field." (Hat tip to zimmer at Patently-O for the quote.) It appears that isolating cDNA clones looks equally easy to Examiners, APJs, and Federal Circuit judges when they fail to consider the underlying facts. Neither is a good idea, nor good for the country.

Leave a comment